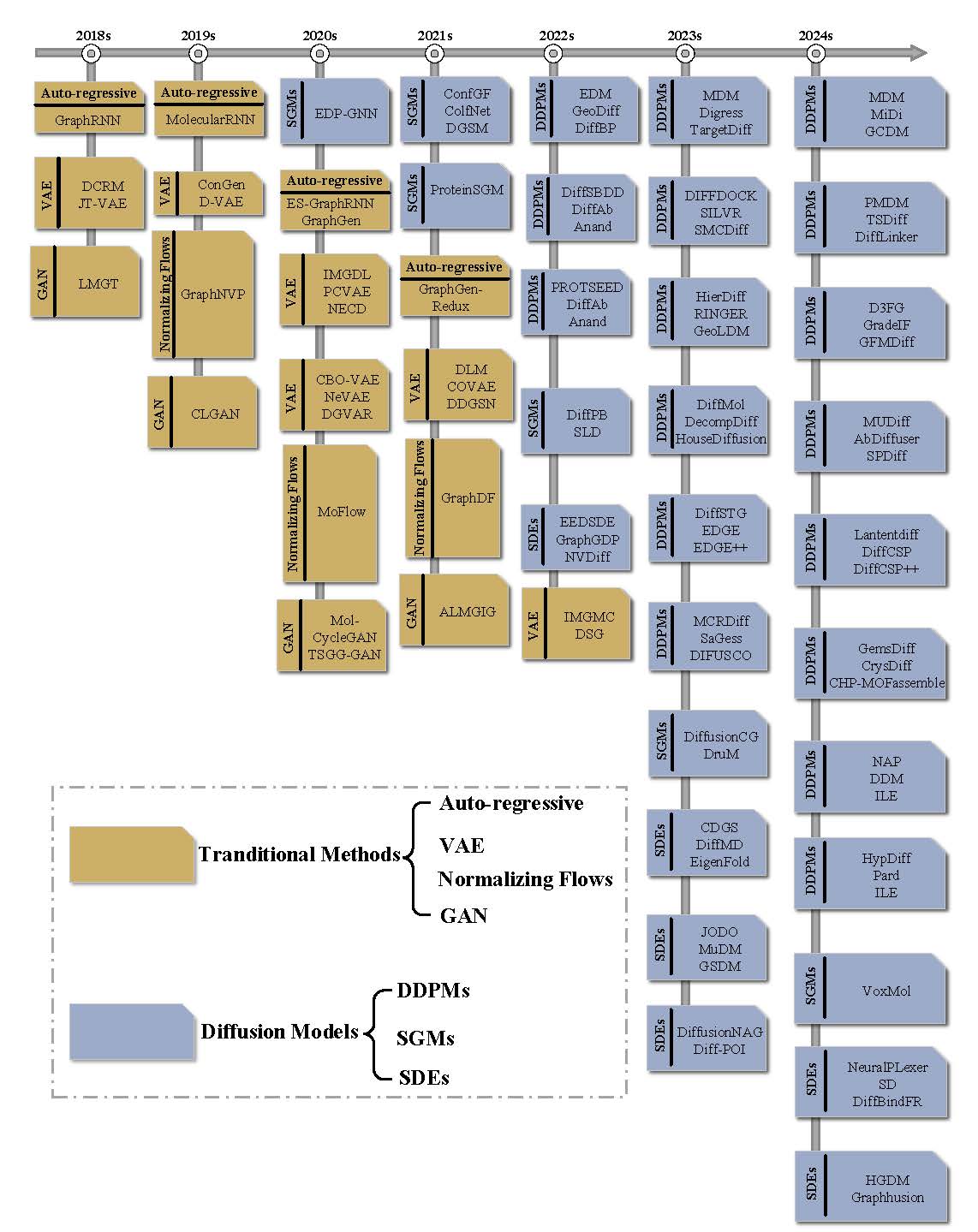
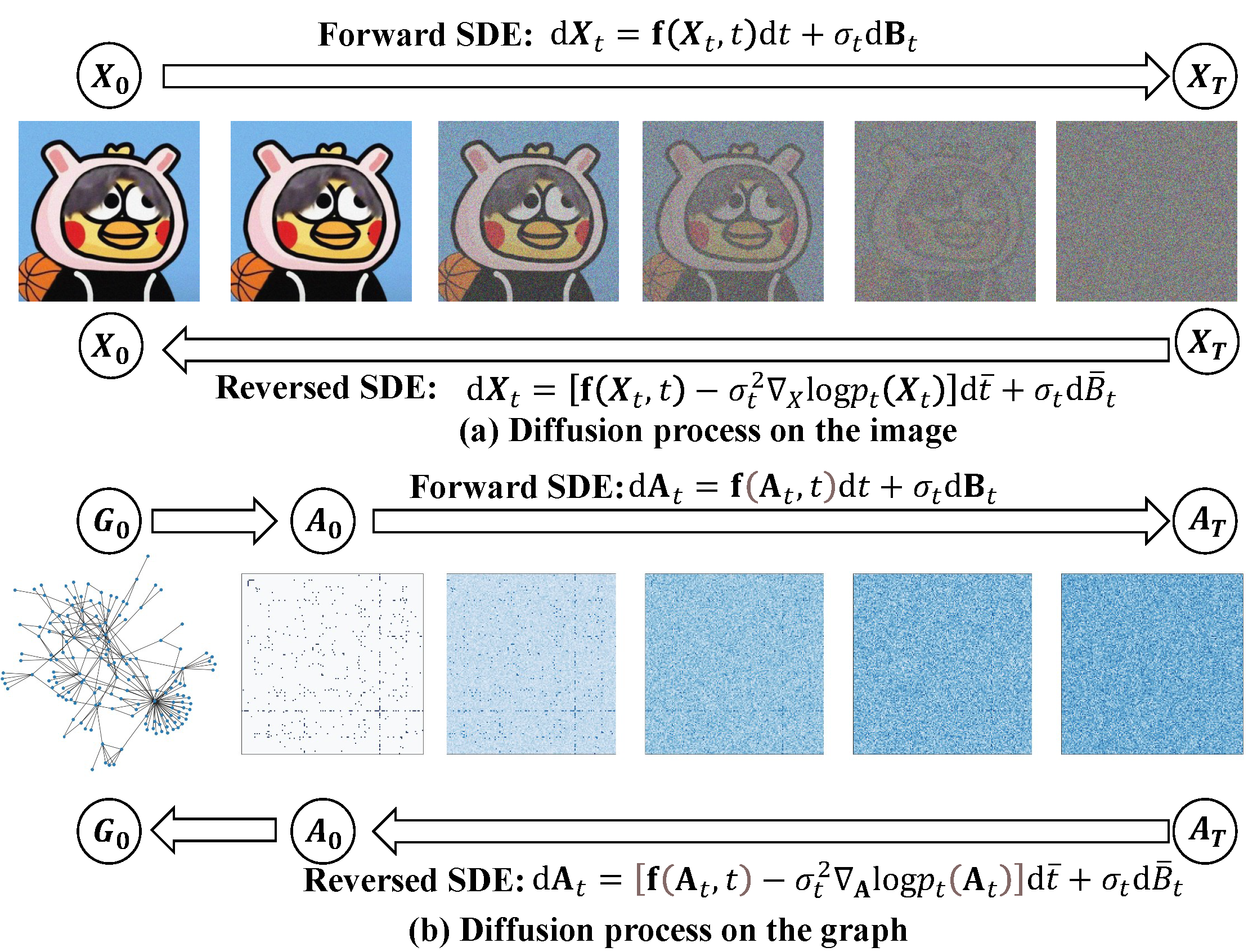
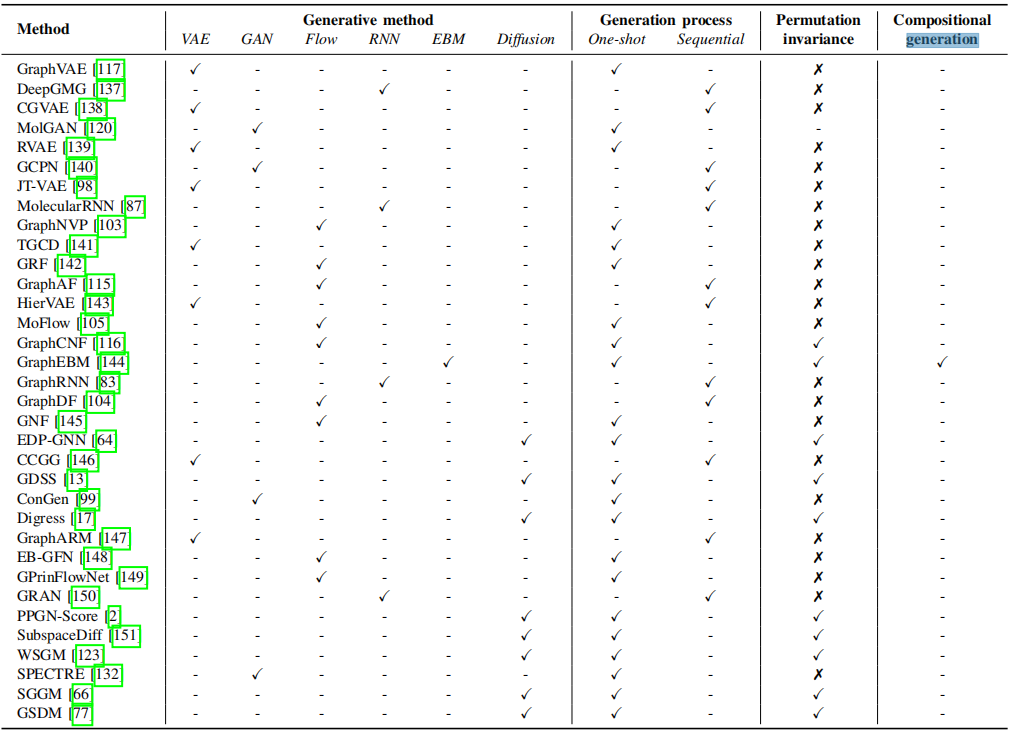

This is the summation of all the methods, datasets, and other survey mentioned in our survey ‘Graph Diffusion Models: A Comprehensive Survey of Methods and Applications’ 🔥. Any problems, please contact shouyuntao@stu.xjtu.edu.cn. Any other interesting papers or codes are welcome. If you find this repository useful to your research or work, it is really appreciated to star this repository ❤️.
| Methods | Paper | Code | Methods | Paper | Code |
|---|---|---|---|---|---|
| DiGress (ICLR-23) | [paper] | [code] | MiDi (ICLR-23) | [paper] | [code] |
| CDGS (NeurIPS-22) | [paper] | [code] | GCDM (ICLR-23) | [paper] | [code] |
| EDM (ICML-22) | [paper] | [code] | Wu et al. (NeurIPS-22) | [paper] | – |
| MDM (AAAI-23) | [paper] | [code] | DiffLinker | [paper] | [code] |
| JODO | [paper] | [code] | SILVR | [paper] | – |
| HierDiff (ICML-23) | [paper] | [code] | – | – | – |
| BIMODAL | [paper] | [code] | RationaleRL (ICML20) | [paper] | [code] |
| GEOLDM (ICML-23) | [paper] | [code] | MGM | [paper] | – |
| LFM AISTATS-20 | [paper] | [code] | RetMol | [paper] | [code] |
| MolGPT | [paper] | – | Bridge (NeurIPS-2022) | [paper] | – |
| Bresson et al. | [paper] | – | FLAG (ICLR-23) | [paper] | [code] |
| LIMO | [paper] | [code] | D3FG NeurIPS-24 | [paper] | [code] |
| Methods | Paper | Code | Methods | Paper | Code |
|---|---|---|---|---|---|
| ConfGF (ICML-21) | [paper] | [code] | DGSM (NeurIPS-21) | [paper] | – |
| GeoDiff (ICLR-22) | [paper] | [code] | ColfNet (ICML-22) | [paper] | – |
| Torsion Diffusion (NeurIPS-22) | [paper] | [code] | DiffMD (AAAI-23) | [paper] | – |
| RINGER | [paper] | – | – | – | – |
| Methods | Paper | Code | Methods | Paper | Code |
|---|---|---|---|---|---|
| DiffBP | [paper] | – | DiffSBDD | [paper] | – |
| TargetDiff (ICLR-23) | [paper] | – | PMDM | [paper] | – |
| D3FG | [paper] | – | – | – | – |
| Methods | Paper | Code |
|---|---|---|
| DIFFDOCK (NeurIPS-22) | [paper] | – |
| EDM-Dock (JCIM) | [paper] | [code] |
| DPL | [paper] | [code] |
| NeuralPLexer | [paper] | – |
| E3BIND (ICLR-23) | [paper] | – |
| Methods | Paper | Code | Methods | Paper | Code |
|---|---|---|---|---|---|
| DiffAb (NeurIPS-22) | [paper] | – | Anand | [paper] | – |
| PROTSEED (ICLR-23) | [paper] | – | ProteinSGM | [paper] | [code] |
| SMCDiff (ICLR-23) | [paper] | [code] | GraDe-IF | [paper] | – |
| EigenFold | [paper] | [code] | – | – | – |
| CGM (NeurIPS-19) | [paper] | [code] | SeqDesign | [paper] | [code] |
| Fold2Seq (ICML-21) | [paper] | [code] | EvoDiff | [paper] | – |
| GVP (ICLR-21) | [paper] | [code] | ESM (NeurIPS-21) | [paper] | – |
| Methods | Paper | Code | Homepage | Methods | Paper | Code | Homepage |
|---|---|---|---|---|---|---|---|
| MotionDiffuse | [paper] | [code] | [homepage] | Modiff | [paper] | – | – |
| Ren et al. (ICASSP-23) | [paper] | – | – | FLAME (AAAI-23) | [paper] | – | – |
| MoFusion (CVPR-23) | [paper] | – | [homepage] | MDM (ICLR-23) | [paper] | [code] | [homepage] |
| MLD (CVPR-23) | [paper] | [code] | [homepage] | PriorMDM | [paper] | [code] | [homepage] |
| Alexanderson et al. (ACM Trans. Graph.) | [paper] | – | – | EDGE (CVPR-23) | [paper] | [code] | [homepage] |
| SceneDiffuser | [paper] | [code] | [homepage] | MoDi (CVPR-23) | [paper] | [code] | [homepage] |
| BiGraphDiff | [paper] | – | – | DiffuPose | [paper] | – | – |
| Methods | Paper | Code | Homepage |
|---|---|---|---|
| Ahn et al. (ICRA-23) | [paper] | [code] | [homepage] |
| HumanMAC | [paper] | [code] | [homepage] |
| TCD (ICRA-23) | [paper] | [code] | – |
| DiffMotion | [paper] | – | – |
| Methods | Paper | Code | Homepage | Methods | Paper | Code | Homepage |
|---|---|---|---|---|---|---|---|
| EDP-GNN | [paper] | – | – | GSDM | [paper] | – | – |
| NVDiff | [paper] | – | – | DPM-GSP | [paper] | – | – |
| DiffSTG | [paper] | – | – | DIFUSCO | [paper] | – | – |
| GraphGDP | [paper] | [code] | – | HouseDiffusion (CVPR-23) | [paper] | [code] | [homepage] |
| NAP | [paper] | – | [homepage] | EDGE | [paper] | – | – |
| DruM | [paper] | – | – | DDM | [Paper] | – | – |
| DiffusionNAG | [paper] | – | – | TSDiff | [paper] | – | – |
| GraphArm | [paper] | – | – | HGDM | [paper] | [code] | – |
| Lee et al. | [paper] | – | – | SaGess | [paper] | – | – |
| SLD | [paepr] | – | – | Diff-POI | [paper] | – | – |
| Brain Diffuser | [paper] | – | – | Lu et al. | [paper] | – | – |
| GBD | [paper] | [code] | – | DiffGraph | [paper] | [code] | – |
| ProGDM | [paper] | [code] | – | R-ode | [paper] | – | – |
| Dataset | Dimensionality | Category | No.of Graphs (G) | No. of Nodes (N) |
|---|---|---|---|---|
| Community-small | 2D | Social | 100 | 11 < N < 20 |
| Ego-small | 2D | Social | 200 | 3 < N < 18 |
| Grid | 2D | Grid | 100 | N <= 400 |
| QM9 | 3D | Bioinformatics/Molecular | 130,831 | 3 < N < 29 |
| ZINC250K | 3D | Bioinformatics/Molecular | 249,456 | 6 < N < 38 |
| Enzymes | 3D | Bioinformatics/Protein | 600 | 9 < N < 125 |
| SBM-27 | 2D | Social | 200 | 24 < N < 27 |
| Planar-60 | 2D | Social | 200 | N = 60 |
| AIDS | 2D | Bioinformatics/Molecular | 2000 | – |
| Synthie | 2D | Social | 300 | N = 100 |
| Proteins | 3D | Bioinformatics/Protein | 1113 | N = 39.1 |
| Methods | Paper | Source | Methods | Paper | Source |
|---|---|---|---|---|---|
| Zinc | [paper] | [source] | GEOM-QM9 | [paper] | [source] |
| GEOM-Drugs | [paper] | [source] | CrossDocked2020 | [paper] | [source] |
| BioLiP | [paper] | [source] | PDBBind | [paper] | [source] |
| SAbDab | [paper] | [source] | – | – | – |
| Methods | Paper | Source | Methods | Paper | Source |
|---|---|---|---|---|---|
| Human3.6M | [paper] | [source] | HumanEva-I | [paper] | [source] |
| HumanAct12 | [paper] | [source] | HumanML3D | [paper] | [source] |
| KIT | [paper] | [source] | BABEL | [paper] | [source] |
| UESTC | [paper] | [source] | 3DPW | [paper] | [source] |
| NTU RGB+D | [paper] | [source] | AIST++ | [paper] | [source] |
| TSG | [paper] | [source] | ZeroEGGS | [paper] | [source] |
| Paper | Url | Source |
|---|---|---|
| Diffusion-based Graph Generative Methods | [paper] | [source] |
| Diffusion Models: A Comprehensive Survey of Methods and Applications | [paper] | [source] |
| A Survey on Generative Diffusion Model | [paper] | [source] |
| Generative Diffusion Models on Graphs: Methods and Applications | [paper] | – |
| A Survey on Graph Diffusion Models: Generative AI in Science for Molecule, Protein and Material | [paper] | – |
| Graph-based Molecular Representation Learning | [paper] | – |
| Generative Models as an Emerging Paradigm in the Chemical Sciences | [paper] | – |
| A Survey on Deep Graph Generation: Methods and Applications (LoG-22) | [paper] | – |
| A Survey on Temporal Graph Representation Learning and Generative Modeling | [paper] | – |
| Human motion modeling with deep learning: A survey | [paper] | – |
| MolGenSurvey: A Systematic Survey in Machine Learning Models for Molecule Design | [paper] | – |
Thanks to Diffusion-based-Graph-Generative-Methods.
 https://github.com/yuntaoshou/Graph-Diffusion-Models-A-Comprehensive-Survey-of-Methods-and-Applications
https://github.com/yuntaoshou/Graph-Diffusion-Models-A-Comprehensive-Survey-of-Methods-and-Applications


Leave a Reply